225x Filetype PDF File size 2.55 MB Source: publications-and-posters.s3.amazonaws.com
A method for repeated neural circuit identification in noisy brain graph data
Elizabeth P. Reilly, Morgan V. Schuyler, Jordan K. Matelsky, William R. Gray-Roncal
The Johns Hopkins University Applied Physics Laboratory
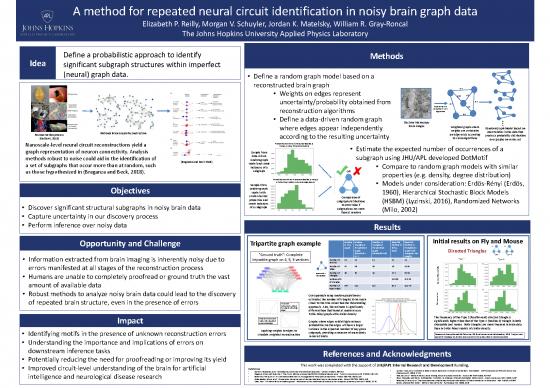
Idea Define a probabilistic approach to identify Methods
significant subgraph structures within imperfect
(neural) graph data. • Define a random graph model based on a
reconstructed brain graph
• Weights on edges represent 0.6
0.95 0.5
uncertainty/probability obtained from 0.3
0.2
Reconstruction 0.9
Computer Vision
reconstruction algorithms Algorithms
0.8
• Define a data-driven random graph Electron Microscopy …
where edges appear independently brain images Weighted graph where Random Graph Model based on
Notional Brain Graph Reconstruction weights are certainties uncertainties in the data that
Reconstruction process according to the resulting uncertainty an edge exists according creates a probability distribution
(Kasthuri, 2015) to vision algorithms over graphs on vertex set
Nanoscale-level neural circuit reconstructions yield a Probability distribution of directed (feedback)
triangle in noisy Drosophila medulla • Estimate the expected number of occurrences of a
graph representation of neuron connectivity. Analysis Sample from
methods robust to noise could aid in the identification of data-driven subgraph using JHU/APL developed DotMotif
(Braganza and Beck 2018) random graph
a set of subgraphs that occur more than at random, such model and count
instances of a • Compare to random graph models with similar
as those hypothesized in (Braganza and Beck, 2018). subgraph OR properties (e.g. density, degree distribution)
Probability distribution of directed (feedback) triangle
in Erdös-Rényi model with similar edge density • Models under consideration: Erdös-Rényi (Erdös,
Sample from
random graph
Objectives model with 1960), Hierarchical Stochastic Block Models
similar desired Comparison of
properties and subgraph distributions (HSBM) (Lyzinski, 2016), Randomized Networks
count instances to determine if
• Discover significant structural subgraphs in noisy brain data of a subgraph subgraph occurs more (Milo, 2002)
• Capture uncertainty in our discovery process than at random
• Perform inference over noisy data Results
Number Number Numberof Expected Expected Initial results on Fly and Mouse
Opportunity and Challenge Tripartite graph example in True Triangles in Triangles in Number in Number in
Graph Thresholded Thresholded Erdos- Probabilistic
Graph Graph Reyni Graph with Directed Triangles
“Ground truth”: Complete (threshold = (threshold=0.8) Triangular dist Type 1 Type 2
0.7) weights
• Information extracted from brain imaging is inherently noisy due to tripartite graph on 4, 5, 3 vertices Number of 12 12 12 12 12 Type 1 Type 2
Vertices a
Number of 47 46 36 33 42.95 l
l
errors manifested at all stages of the reconstruction process Edges u
d
e
Number of 60 39 24 27.5 59.58 m
Open triad. 1
a
l
missing edge Triangles i
h
• Humans are unable to completely proofread or ground truth the vast p
Variance of # 116.88 156.29 o
s
of triangles o
Dr
amount of available data Numberof 145 122 115 82.5 111.76
Open Triads
a
n
i
• Robust methods to analyze noisy brain data could lead to the discovery Our approach using random graph theory t
e
r
estimates the number of triangles to be much e
s
closer to the true value than the thresholding u
of repeated brain structure, even in the presence of errors Probability Mo
Distribution Function approach. Also, the estimate is significantly
for weights of existing
edges different than that found at random in an
Erdös-Rényi graph with similar density. The frequency of the Type 2 (feedforward) directed triangle is
Probability Distribution
Impact Function for weights of Graphs where edges exhibit high variance significantly higher than that of the Type 1 (feedback) triangle in both
non-existent edges probabilities on the edges will have a larger drosophila and mouse. Both triangles are more frequent in brain data
Applying weights to edges to variance in the expected number of any given than in Erdös-Rényi models of similar density.
• Identifying motifs in the presence of unknown reconstruction errors simulate weighted reconstruction subgraph, providing a measure of uncertainty Datasets are Drosophila medulla (Takemura, 2013) and mouse retina (Helmstaedter, 2013). Weights were
• Understanding the importance and implications of errors on in our estimate. applied to the edges to simulate noisy reconstruction as described in tripartite example.
downstream inference tasks References and Acknowledgments
• Potentially reducing the need for proofreading or improving its yield
This work was completed with the support of JHU/APL Internal Research and Development Funding.
• Improved circuit-level understanding of the brain for artificial References • Lyzinski, Vince, et al. "Community detection and classification in hierarchical stochastic blockmodels." IEEE Transactions on Network Science and
• Kasthuri, Narayanan, et al. "Saturated reconstruction of a volume of neocortex." Cell 162.3 (2015): 648-661. Engineering 4.1 (2016): 13-26.
• Braganza, Oliver, and Heinz Beck. "The Circuit Motif as a Conceptual Tool for Multilevel Neuroscience." Trends in neurosciences 41.3 (2018): 128-136. • Milo, Ron, et al. "Network motifs: simple building blocks of complex networks." Science 298.5594 (2002): 824-827.
intelligence and neurological disease research • Takemura, Shin-ya, et al. "A visual motion detection circuit suggested by Drosophila connectomics." Nature 500.7461 (2013): 175. • Borst, Alexander, and Moritz Helmstaedter. "Common circuit design in fly and mammalian motion vision." nature neuroscience 18.8 (2015): 1067.
• Helmstaedter, Moritz, et al. "Connectomic reconstruction of the inner plexiform layer in the mouse retina." Nature 500.7461 (2013): 168. • Takemura, Shin-ya, et al. "The comprehensive connectome of a neural substrate for ‘ON’motion detection in Drosophila." Elife 6 (2017): e24394.
• Erdös, Paul. "On the evolution of random graphs." Publications of the mathematical institute of the Hungarian academy of sciences 5 (1960): 17-61. • Sporns, Olaf, and Rolf Kötter. "Motifs in brain networks." PLoS biology 2.11 (2004): e369.
no reviews yet
Please Login to review.